Public defence in Biomedical Engineering, M.Sc. Ashwin Natarajan
When
Where
Event language(s)
Opponent: Professor Tim Liedl, Ludwig Maximilian University of Munich, Germany
Custos: Associate Professor Anton Kuzyk, Aalto University School of Science, Department of Neuroscience and Biomedical Engineering
Defence will be organized on campus and via Zoom: https://aalto.zoom.us/j/64441997272
The doctoral thesis will be publicly displayed 10 days before the defence in the publication archive of Aalto University.
Public defence announcement:
Nucleic acids, deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), are molecules that store and transmit the genetic information in cells of all living organisms, from bacteria to plants to animals. Nucleic acids are not just found in Nature but can also be synthesized chemically. These synthetic nucleic acid molecules can be completely identical to the one found in Nature based on their sequence. Like Legos, the sequences of each nucleic acid strand bind specifically to their complement. By manipulating the nucleic acid sequence, one can control their interactions, thereby programming the nucleic acid strands to bind in a predictable manner with other strands to form desired structures. Such intricate structures formed in nanoscale are called nucleic acid nanostructures.
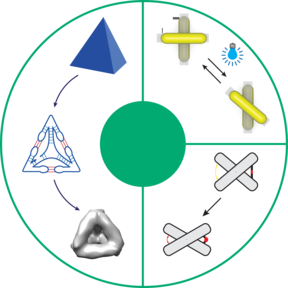
The principles to fabricate these nanostructures have been laid forth in the early 1980s. However, a major leap forward came with the introduction of DNA origami technique, which involves folding a long single strand of DNA called ‘scaffold’ strand using hundreds of short strands called ‘staple’ strands. The short strands are designed to staple parts of the scaffold strand together, thereby routing the scaffold strand to form the prescribed shape. With DNA origami technique, almost arbitrary shapes can be fabricated in nanoscale. With the ability to bind to other functional materials such as proteins, metal nanoparticles, and fluorophores, nucleic acid nanotechnology has become a highly sophisticated nanofabrication technique.
Nucleic acid nanostructures, like any other nanostructure, have to form a user-defined shape, need to exhibit functionalities such as responding to certain external stimulus, and showcase that they can be applied to solve research questions. In this thesis, we demonstrate a general design pipeline for RNA-based nanostructures, a stimuli responsive DNA origami-based reconfigurable nanostructure that responds to light not directly but through the pH changes caused by a photo-responsive medium, and an application for the DNA origami-based nanostructure to study the structural changes induced by a protein that is essential in regulating the activity of most genes.
The thesis highlights the versatility of nucleic acids as a nanoscale building material, and how the fabricated nanostructures could pave the way toward complex nanostructures with tailored functionalities in biophysics, bioimaging, DNA-based nanomachines, and smart nanomaterials.
Contact details of the doctoral student: ashwin.natarajan@aalto.fi, +358503211931






